Detailed Running
Code Structure
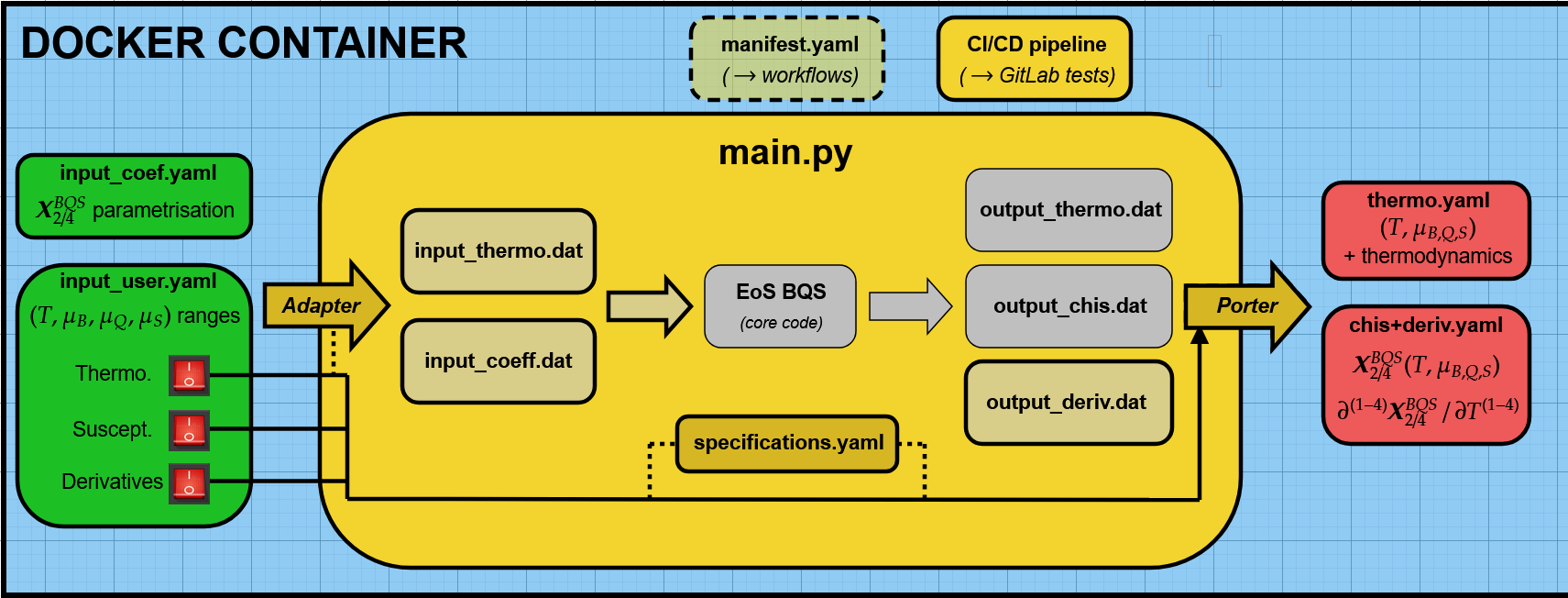
This module performs the computation of thermodynamic and susceptibility data based on the EoS BQS (Equation of State for Baryon, Charge, and Strangeness) framework. The workflow is implemented within a Docker container, ensuring portability and consistency. The process involves parsing input parameters, performing core computations, and generating output files for further analysis. Below is a step-by-step explanation:
Input Files: - input_coef.yaml: Defines the parametrization for \(X^{\text{BQS}}_{2/4}\) coefficients. - input_user.yaml: Specifies the ranges for \(T\), \(\mu_B\), \(\mu_Q\), and \(\mu_S\) as well as user-defined options for:
Thermo.: Thermodynamic data
Suscept.: Susceptibility data
Derivatives: Temperature derivatives of thermodynamic functions
Preprocessing via Adapter: The adapter processes the input YAML files (input_coef.yaml and input_user.yaml) and generates intermediate input files in .dat format: - input_thermo.dat: For thermodynamic computations - input_coeff.dat: For coefficient-based parametrizations
Core Computation: The main script main.py handles the execution of the EoS BQS core code. This computation utilizes the .dat files generated in the preprocessing step to calculate: - Thermodynamic data - Susceptibilities - Temperature derivatives
Output Files: The results of the computation are saved in the following files: - output_thermo.dat: Contains computed thermodynamic data (\(T\), \(\mu_B\), \(\mu_Q\), \(\mu_S\) and related values). - output_chis.dat: Includes susceptibility data, \(\chi\). - output_deriv.dat: Outputs temperature derivatives of \(X^{\text{BQS}}_{2/4}\).
Postprocessing via Porter: A postprocessing step handled by “Porter” converts the output .dat files into YAML format for better compatibility and downstream workflows: - thermo.yaml: YAML representation of thermodynamic data. - chis+deriv.yaml: YAML representation of susceptibility and temperature derivative data.
Specifications: Additional configurations and specifications are provided in specifications.yaml, which ensures consistency and reproducibility across different setups.
CI/CD Integration: The workflow includes a manifest.yaml file for integration with CI/CD pipelines (e.g., GitLab tests), ensuring automated testing and validation.